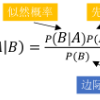
Familial DNA search evaluates the genetic relatedness of two individuals by comparing the likelihood of their observed DNA profiles under two competing hypotheses-the null hypothesis that the individuals are unrelated and the alternative hypothesis that they are related-most commonly through the likelihood ratio (LR). Standard LR-based approaches typically assume a uniform genetic background; however, this assumption is rarely valid due to population substructure, where allele frequencies vary among subpopulations and can bias relationship inference. Existing modifications-such as LR calculations based on average allele frequencies (LRLAF) and strategies using maximum, minimum, or average likelihood ratios (LRMAX, LRMIN, LRAVG)-help mitigate these challenges but remain limited in their ability to fully address subpopulation differences. This study introduces a new LR-based statistic, LRCLASS, which incorporates a classification step using the Naive Bayes classifier to account for nuisance parameters associated with unknown subpopulation origins. In LRCLASS, the two DNA profiles being compared are jointly assigned to a subpopulation group via Naive Bayes before LR computation. Empirical evaluations using Thai population data show that LRCLASS achieves higher statistical power for detecting full-sibling relationships than existing LR-based methods. We further assessed multinomial logistic regression as an alternative classifier and found its performance comparable to that of Naive Bayes, suggesting flexibility in classifier choice. Overall, integrating the Naive Bayes classifier with LR computation offers a robust strategy for addressing population substructure in familial DNA search and highlights the broader potential of combining supervised learning techniques with forensic statistical methodologies to enhance the accuracy and reliability of genetic relationship testing.
翻译:家族DNA搜索通过比较两个个体在两种竞争假设下观察到的DNA谱似然性来评估其遗传相关性——零假设为个体间无亲缘关系,备择假设为个体间存在亲缘关系,通常采用似然比(LR)进行计算。标准的基于LR的方法通常假设统一的遗传背景;然而,由于种群亚结构的存在,这一假设很少成立——等位基因频率在亚群间存在差异,可能导致亲缘关系推断产生偏差。现有改进方法(例如基于平均等位基因频率的LR计算(LRLAF)以及使用最大、最小或平均似然比的策略(LRMAX、LRMIN、LRAVG))有助于缓解这些挑战,但在全面处理亚群差异方面仍存在局限。本研究提出一种新的基于LR的统计量LRCLASS,其通过整合朴素贝叶斯分类器的分类步骤,以处理与未知亚群来源相关的干扰参数。在LRCLASS中,待比对的两种DNA谱在LR计算前通过朴素贝叶斯方法被联合分配至一个亚群组。基于泰国人群数据的实证评估表明,在检测全同胞关系方面,LRCLASS相比现有基于LR的方法具有更高的统计功效。我们进一步评估了多项逻辑回归作为替代分类器的性能,发现其表现与朴素贝叶斯分类器相当,表明分类器选择具有一定灵活性。总体而言,将朴素贝叶斯分类器与LR计算相结合,为处理家族DNA搜索中的种群亚结构提供了稳健策略,并凸显了监督学习技术与法医统计方法结合在提升遗传关系检测准确性与可靠性方面的广阔潜力。