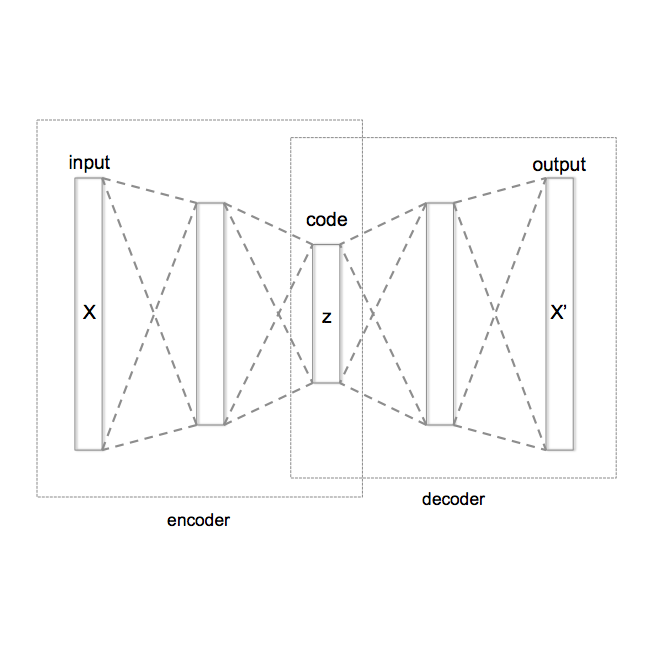
Automatic segmentation of brain abnormalities is challenging, as they vary considerably from one pathology to another. Current methods are supervised and require numerous annotated images for each pathology, a strenuous task. To tackle anatomical variability, Unsupervised Anomaly Detection (UAD) methods are proposed, detecting anomalies as outliers of a healthy model learned using a Variational Autoencoder (VAE). Previous work on UAD adopted a 2D approach, meaning that MRIs are processed as a collection of independent slices. Yet, it does not fully exploit the spatial information contained in MRI. Here, we propose to perform UAD in a 3D fashion and compare 2D and 3D VAEs. As a side contribution, we present a new loss function guarantying a robust training. Learning is performed using a multicentric dataset of healthy brain MRIs, and segmentation performances are estimated on White-Matter Hyperintensities and tumors lesions. Experiments demonstrate the interest of 3D methods which outperform their 2D counterparts.
翻译:大脑异常的自动分解具有挑战性,因为各个病理学和另一个病理学之间差异很大。 目前的方法受到监督,需要为每个病理学提供大量附加说明的图像,这是一项艰巨的任务。 为了应对解剖变异性,提出了无监督异常检测(UAD)方法,发现异常是使用变异自动读数器(VAE)学习的健康模型的异端。UAD以前的工作采用了2D方法,这意味着MRI是作为独立切片的集合处理的。然而,它并没有充分利用MRI中包含的空间信息。在这里,我们提议以3D方式进行UAD,比较2D VAE和3D VAEs。作为侧面贡献,我们提出了一个新的损失函数,用于进行强健健的训练。学习使用多中心的健康脑调数数据集进行,对白发超重度和肿瘤损害的分解性表现进行了估计。 实验显示了3D方法的兴趣,这些方法超越了2D对等。