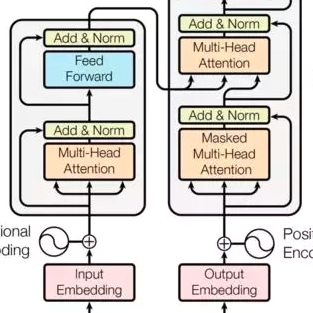
Transformers generate valid and diverse chemical structures, but little is known about the mechanisms that enable these models to capture the rules of molecular representation. We present a mechanistic analysis of autoregressive transformers trained on drug-like small molecules to reveal the computational structure underlying their capabilities across multiple levels of abstraction. We identify computational patterns consistent with low-level syntactic parsing and more abstract chemical validity constraints. Using sparse autoencoders (SAEs), we extract feature dictionaries associated with chemically relevant activation patterns. We validate our findings on downstream tasks and find that mechanistic insights can translate to predictive performance in various practical settings.
翻译:Transformer模型能够生成有效且多样化的化学结构,但其捕捉分子表示规则的内部机制尚不明确。本文对基于药物类小分子训练的自回归Transformer进行机制分析,从多个抽象层次揭示其能力背后的计算结构。我们识别出与低层次句法解析及更抽象的化学有效性约束相一致的计算模式。通过稀疏自编码器(SAEs),我们提取了与化学相关激活模式关联的特征词典。我们在下游任务中验证了这些发现,并证明机制性见解可转化为多种实际场景中的预测性能提升。